learningMatlab
Worked solution
Plotting / Fitting with a model curve
The aim is to plot and fit data to achieve something like the following:
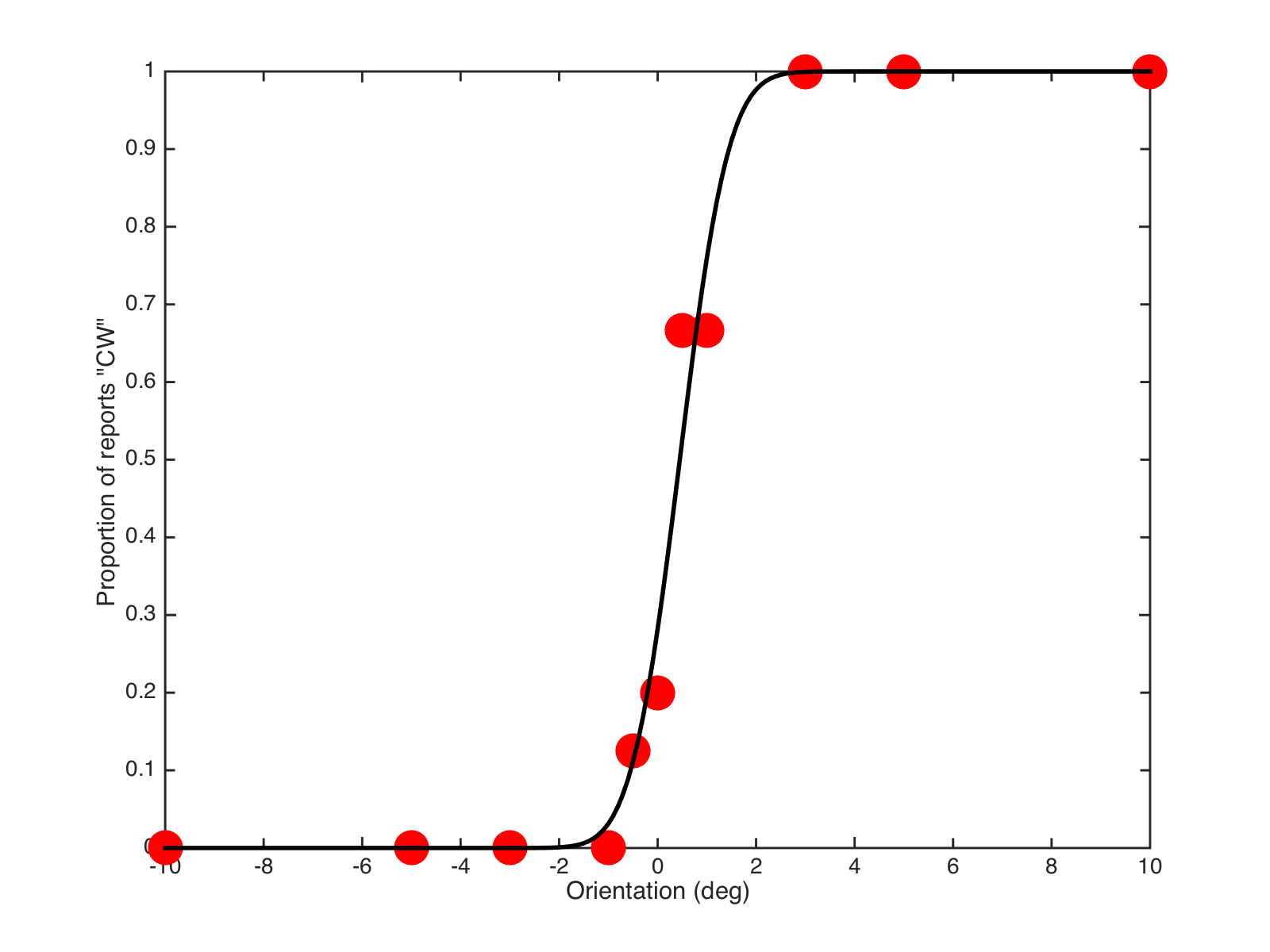
-
The file
summaryData.matcontains data to work with. At the command prompt, do the following:load('summaryData.mat') % will load "data" data % find out what's in it. -
Plot
data, using the numbers in the first column as the x-values, the second column as the y-values.figure plot(data(:,1) , data(:,2), 'o', 'markersize',20, 'markerfacecolor', 'b') xlabel('Orientation (deg)') ylabel('p(CW responses)') % and save a copy for your records % help print % help gcf % ... to see why/how the following line works: print(gcf, 'myFigure.pdf', '-dpdf') -
A “nice” mathematical function / curve, that has such a shape and can be controlled with two convenient parameters is the cumulative version of the normal distribution (or Gaussian curve).
x = -10:0.2:10; % many x values -10....+10 % plot the CUMULATIVE / s-shaped function with mu=0, sigma=3 c = normcdf(x, 0, 3); figure, plot(x, c , 'r', 'linewidth',2); xlabel('x values'); ylabel('y values'); title('normal CDF: \mu=0, \sigma=3'); -
The 0 and 3 in the above example correspond to the mean (µ) and standard deviation (σ) of a normal distribution. Try plotting the CDF curve for σ = 1 (magenta curve) and σ = 5 (cyan) on the same plot. Then try changing µ, say µ = -2.5 (green) and µ = +3.4 (blue). What happens to the curves?
x = -10:0.2:10; % many x values -10....+10 % plot the CUMULATIVE / s-shaped function with other values figure subplot(1,2,1) plot(x, normcdf(x, 0, 3) , 'r', 'linewidth',2); hold on plot(x, normcdf(x, 0, 1) , 'm', 'linewidth',2); plot(x, normcdf(x, 0, 5) , 'c', 'linewidth',2); title('changing \sigma') subplot(1,2,2) plot(x, normcdf(x, 0, 3) , 'r', 'linewidth',2); hold on plot(x, normcdf(x, -2.5, 3) , 'g', 'linewidth',2); plot(x, normcdf(x, +3.4, 3) , 'b', 'linewidth',2); title('changing \mu') -
The fitting problem is: we’d like to know the “best” combination of µ and σ, such that the smooth curve they describe gets as close as possible to the data points. “Close” here means: in such a way that the squared error between the data and the corresponding points on the smooth curve is minimal… the least squares solution.
-
Before we can hand over the fitting to matlab we need to do the following:
-
we need re-wrap the
normcdffunction, so that it accepts the input arguments in a different order. Currently it’s:y = normcdf( x, mu, sigma ), wherexare the values for which you want to find the y-values… but really we want it to bey = myNormcdf(p, x), wherep = [mu, sigma]i.e. the two parameters stuck together in a list / vector. Look at the filemyNormcdf.mfor an example of how to do this. -
we need/may want to calculate the squared error between the data (the data-y-values at each point in x) and the model/fit with a particular combination of mu/sigma (also at the corresponding x-values). Look at the file
sqerror.mfor an example of how to do this. The aim here is to get a single number that summarizes how close our guess of the curve is to the y-values/data. Our guess of the curve is determined by the choice of µ and σ. -
just to check what that allows us to do… we can now ask for e.g.
p=[0, 3.0]what is the error between that curve and the data. Next, what if I update the values top=[1.0, 3.0]… does the error get bigger or smaller? If it gets bigger, we are moving away from a good value forp, if the error gets smaller, then the secondpwas better!% with data loaded from summaryData! e1 = sqerror([0, 3], data(:,1), data(:,2) ) e2 = sqerror([1.0, 3], data(:,1), data(:,2) ) % which is better?? -
the matlab function
lsqcurvefit()will do this calculation internally and - by trying out many, many settings forp, find the value for it that gives the smallest possible error (the least squares solution!). -
So: to put this all together now, do
help lsqcurvefitto have a look at how this works in general, then try:% with data loaded from summaryData! bestP = lsqcurvefit(@myNormcdf, [0, 1], data(:,1), data(:,2)) % the @ in @myNormcdf is needed to make sure matlab knows you are handing it a % function! look at "help function_handle" if you want to know more about this. % the [0, 1] is the value for P at which you want to start the whole process bestP = 0.4501 0.7789 -
now that we have the best choice of
p(stored inbestP), we can use it to calculate the curve at lots of x-values we want:newX = linspace(-10,10, 101); % 101 values between -10 and 10 % and our myNormcdf() function is perfect for calculating these! yFit = myNormcdf(bestP, newX); % now plot the data and fit! figure plot(data(:,1) , data(:,2), 'o', 'markersize',20, 'markerfacecolor', 'b') xlabel('Orientation (deg)') ylabel('p(CW responses)') hold on plot(newX, yFit, 'k-', 'linewidth',2) -
Et voilà